Part:BBa_K3264024
TALEsp-vioA-vioB-vioC-vioE
This part contains deoxyviolacein expression genes and TALE stabilized promoter sp2. The complete pathway of biosynthesis of deoxyviolacein, a light purple pigment Despite simply laying a complete database of spideroin, GreatBay_SZ this year also approached one major industry where spider silk held great potential: the cloth industry. Identifying the current chemical compound pollution during dying process as well as the damage brought by chemical fiber itself, we realized that the typically non-environmentally friendly material of cloth manufacture can be replaced by spider silk, as thread to weaved the cloth, and natural dyes, as pigments that granted cloth color.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1513
Illegal NheI site found at 1921
Illegal NheI site found at 4184 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 113
Illegal XhoI site found at 2152
Illegal XhoI site found at 2692 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 6966
Illegal AgeI site found at 747
Illegal AgeI site found at 849
Illegal AgeI site found at 1767
Illegal AgeI site found at 2277
Illegal AgeI site found at 7162 - 1000COMPATIBLE WITH RFC[1000]
Usage in biology
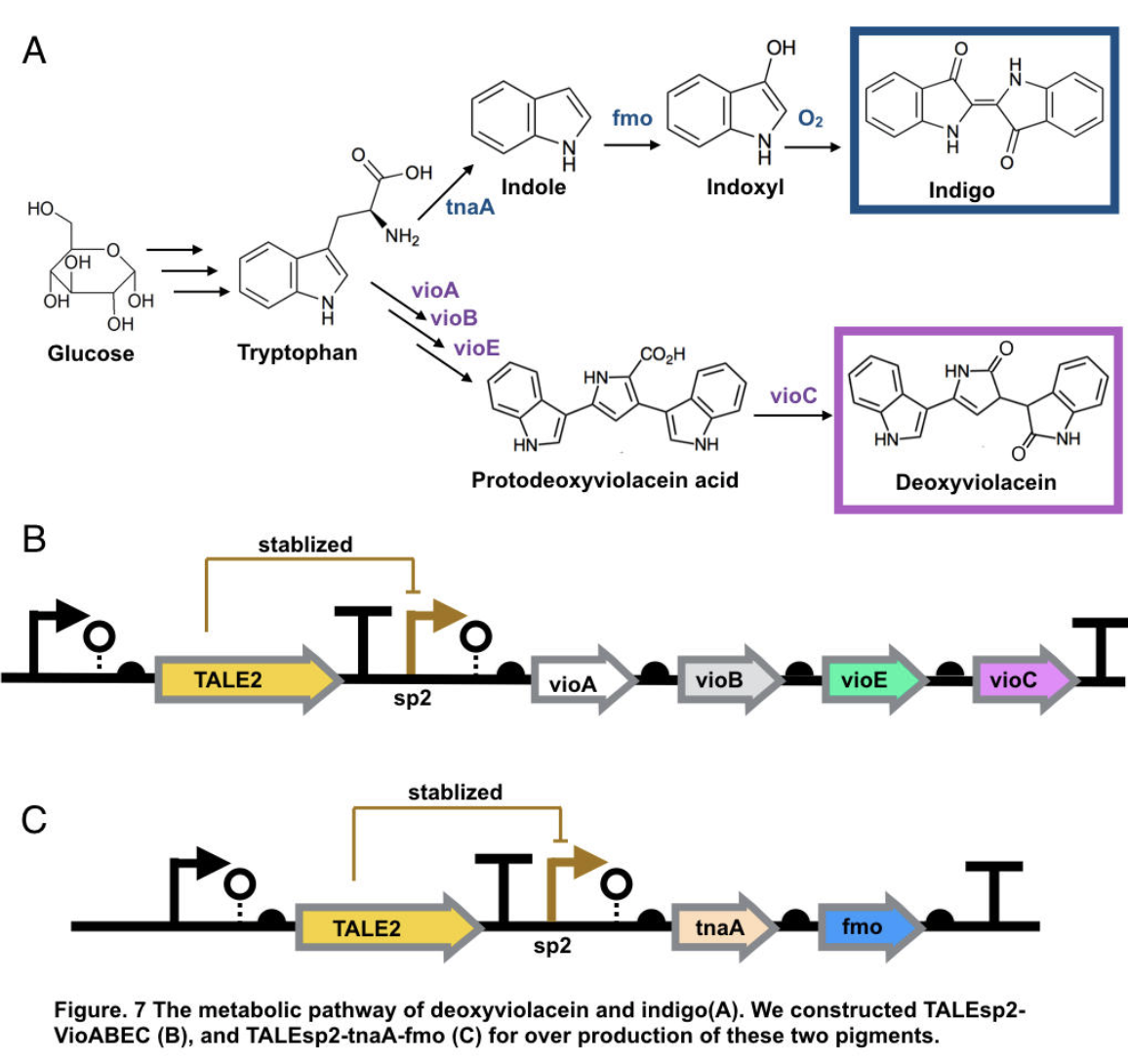
The pigment deoxyviolacein is produced from L-tryptophan in E.coli via a pathway involving four enzymes VioA, VioB, VioE, VioC, only vioD is excluded through the pathway. GreatBay_SZ 2019 borrow from team SHSBNU_China to acquire part thsR- BBa_K274003, which include one thiosulfate sensor and vioABDE which synthesize proviolacein. As shown in the graph below, different arrangement of VioA-E can synthesize pigment with deviate color. Our team wants to substitute VioD to vioC that gives as a light pink/purple pigment
Aiming for stable and high efficient production through deoxyviolacein pathway, using the original thiosulfate sensor cannot yield us high concentration of pigment. Instead, we look into GreatBay_China_2018’s stabilized promoter BBa_K2753019, Talesp2 from the pTale family. Transcription-activator-like-effector (TALE) stabilised promoters are a type of promoters able to untie gene expression level from gene copy number using an incoherent feed forward loop (iFFL) in which transcription-activator-like effectors (TALEs) function as a perfectly non-cooperative negative regulation. While copy number accretes gene expression, it also elevates the repression to the gene expression, thus has canceled out the effect of copy number on expression level. Thus using Talesp2, we comprehend that it can yield high quantitative results
Indigo, though without much significant medical usage, is the most classic ancient dye uniquely capable of producing the signature tones in blue denim. The current production of indigo not only release hazardous chemical compound, but also requires an excess, usually toxic, reducing agent to reduce the insoluble indigo into soluble leucoindigo. Replacement of reducing agent were identified in the past, yet none established as fast nor cost effective. We therefore researched an alternative indigo production pathway. With L-tryptophan, indole is formed by enzyme tnaA in E.coli. In the presence of indole and oxygen, FMO catalyzes the addition of a hydroxyl group to indole generating the intermediate indoxyl that can spontaneously oxidize to form indigo.
Characterization
To achieve over production of both pigments, we utilized the stabilized promoter BBa_K2753019 from GreatBay_China(2018), transcription-activator-like-effector (TALE) stabilised promoters that untie gene expression level from gene copy number. We designed to place TALEsp2 promoter on standard pSC101 backbones. Combining the pathway with promoter, we hence characterized and obtained pigments by constructing part TALEsp2-VioABEC for deoxyviolacein through BBa_K2753019, BBa_K726015, and a new basic part BBa_K3264008, a gene-transcript of VioC. Meanwhile, we also constructed a new composite part TALEsp2-tnaA-FMO for indigo.
Deoxyviolacein has long be identified as secondary metabolity actively against pathogenic bacteria like Pseudomonas aeruginosa and Staphylococcus aureus, and leukemia, lung cancer, human uveal melanoma, and lymphoma cells[10,11]. It also served other purpose like natural pigments. The importance of violacein urged us to search for over production of both metabolities. The hidden pathway for production is encoded by the VioABCDE operon. Bio-synthesis starts from L-tryptophan, converted into protodeoxyviolaceinic acid by VioA, VioB and VioE enzymes, and then into deoxyviolacein is therefore produced with the activation of VioC gene[10]. All promoters are placed on the standard biobrick assembly compatible backbone pSC101. We reassembled plasmid thsR- BBa_K274003 by PCR vioAB, VioE and talesp2, only VioC was synthesized de novo by Genescript. All parts were assembled using Gibson Assembly.
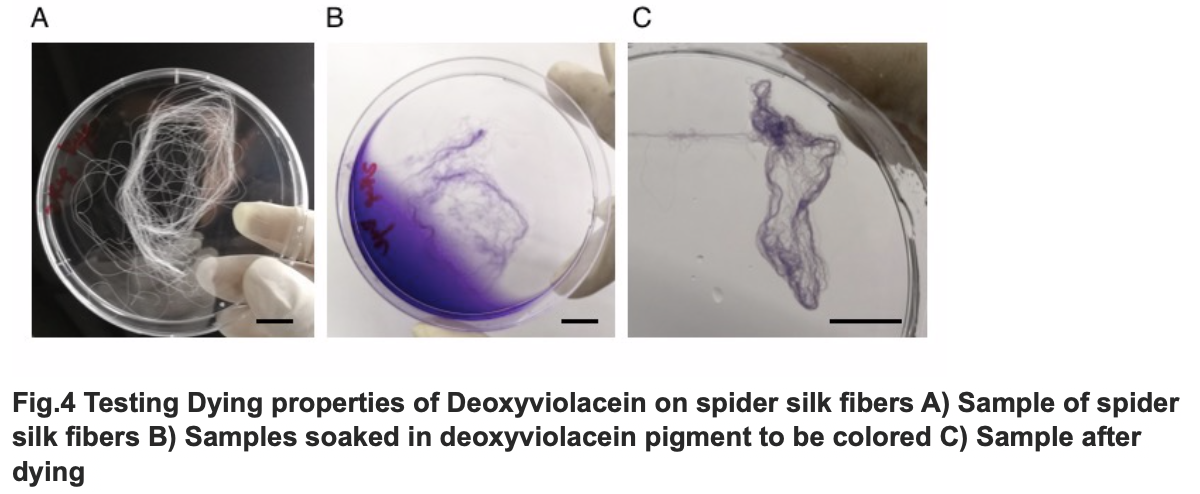
After we acquire pure extract deoxyviolacein pigment, we test it dying properties upon our spider silk sample. As documented as the process below, the pigment can be well soaked into fibers to give it light purple color.
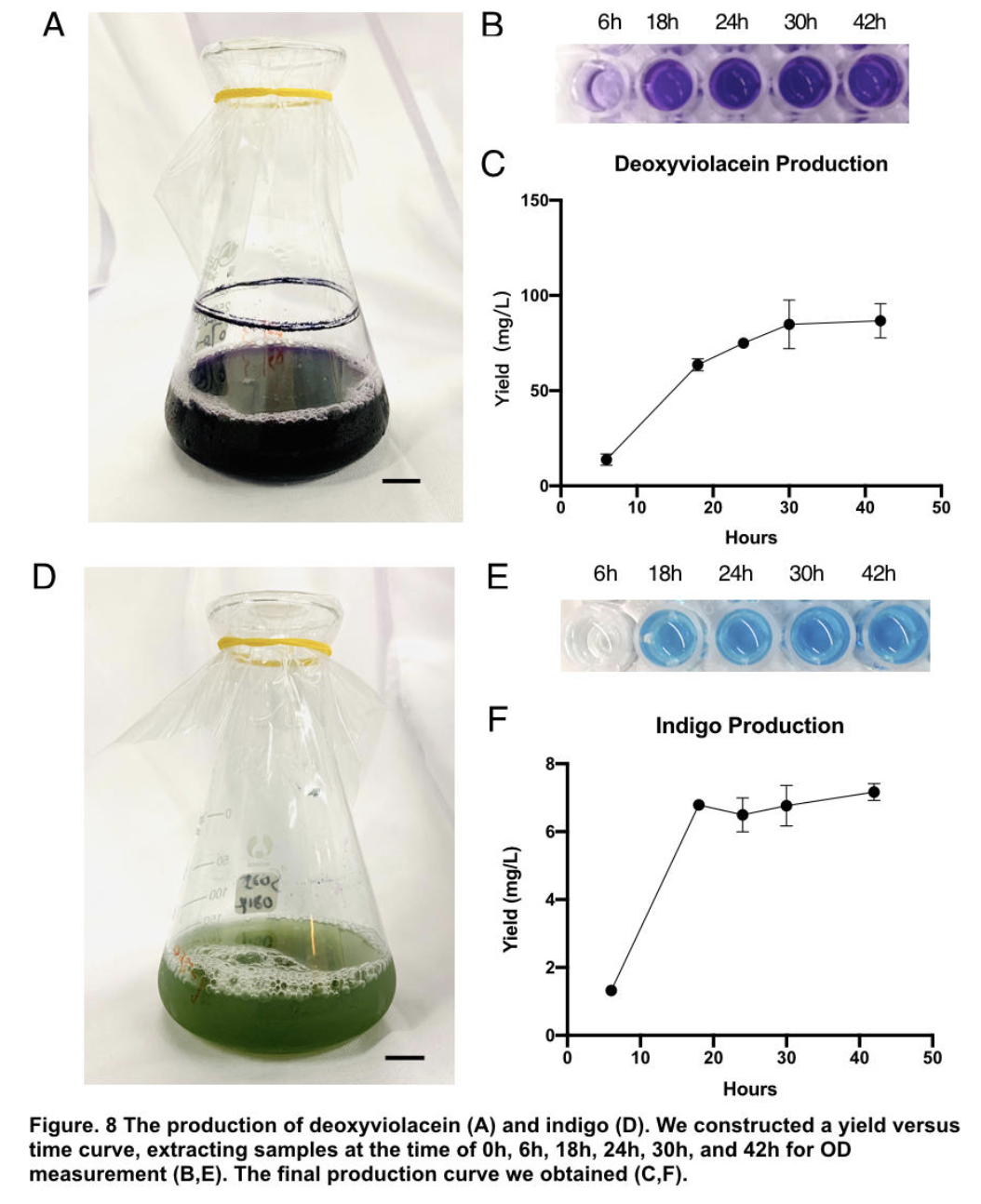
The influence of talesp2 on the production and accumulation of deoxyviolacein is remarkable, with the highest yield round 90~100mg/L which nearly matched the standard of pure extracts deoxyviolacein. It can be concluded that talesp2 has positive influences on deoxyviolacein’s metabolic reaction, which it stabilized towards an optimal level of gene expression that produce just enough enzyme to metabolize the substrate. After we acquire pure extract deoxyviolacein pigment, we test it dying properties upon our spider silk sample. As documented as the process below, the pigment can be well soaked into fibers to give it light purple color.
Pigments were obtained by extracting pigments after 42 hours of shake-flask incubation (without iptg) using solvent ethanol for violacein, and DMSO for indigo respectively. Through calculation based on standard indigo product and OD measurement of oh, 6h, 18h, 24h, 30h, 42h (peak of production), we are able to construct a yield versus time curve. Through that, we concluded our deoxyviolacein yields 85.81±9.09mg/L maximum and indigo yields 6.97±0.44mg/L maximum.
Testing Dying Properties on Spider silk Fibers
Reference
[1]Rodrigues, André L., et al. “Systems Metabolic Engineering of Escherichia Coli for Production of the Antitumor Drugs Violacein and Deoxyviolacein.” Metabolic Engineering, vol. 20, 2013, pp. 29–41., doi:10.1016/j.ymben.2013.08.004. [2]Wen, Rui, et al. “Molecular Cloning and Analysis of the Full-Length Aciniform Spidroin Gene from Araneus Ventricosus.” International Journal of Biological Macromolecules, vol. 117, 2018, pp. 1352–1360., doi:10.1016/j.ijbiomac.2017.12.090.
| None |